Abstract
Introduction Extramedullary disease (EMD) is seen in up to 20% of multiple myeloma (MM). It is associated with poor outcomes despite the use of autologous stem cell transplant (ASCT) and directed agents, however, the biological mechanisms driving this phenotype are poorly understood. In this study, we explored the clinical and genomic characteristics of EMD using the CoMMpass registry and a well-defined cohort at our institute.
Methods The CoMMpass study is a prospective, international registry of newly diagnosed MM. A total of 1143 patients had clinical data. Patients were classified as EMD if they developed an EMD at any point. Genomic data from bone marrow (BM) samples at diagnosis (whole genome sequencing, whole exome sequencing (WES), and RNA sequencing (RNAseq) were compared between EMD and non-EMD. A multivariable model was constructed based on univariable results. Differential expression (DEA) and gene set enrichment analysis (GSEA) were performed after normalization using Hallmark and MM transcriptomic subgroups. The top 24 most commonly mutated genes in MM were selected. A cohort of 67 EMD cases and 81 non-EMD cases diagnosed within the same period was constructed from the Weill Cornell (WCM) MM registry. Diagnosis of MM was confirmed using 2014 IMWG criteria. In preparation for multiplex immunofluorescence staining and digital image analysis (MxIF) visual assessment of single marker screening immunohistochemistry (IHC) was performed on a pilot cohort (n=5) of matched BM and EMD WCM samples using antibodies from the developed MxIF panel (CD3, CD8, FOXP3, CD56, CD138, CD68), and correlated with concurrent flow cytometry (MFC) data; in addition, WES and RNAseq was also performed in matched BM and EMD samples
Results EMD patients presented with lower median M-protein and higher hemoglobin (Hb). The median follow-up for the cohort was 5.4 years after diagnosis. The median overall survival (mOS) and progression free survival (PFS) was shorter for EMD. The presence of EMD, anemia at diagnosis, age >65 years at diagnosis, ISS stage, and use of ASCT at the first line were independently associated with worse mOS and PFS. EMD remained associated with worse mOS (HR 1.7, 95% CI 1.3 - 2.2 p <0.001) and PFS (HR 1.5, 95% CI 1.23 - 1.8, p <0.001) in the multivariable model (Table 1). Results did not vary when landmark analysis was used. Although the use of maintenance, ASCT, and triplet induction was associated with improved mOS and PFS, patients with EMD still experienced inferior outcomes when compared to non-EMD.
We did not find statistically significant differences in the frequency of translocations or copy number abnormalities. TP53 mutations and TP53 bi-allelic inactivation (mutation and deletion) were more common in EMD (9.6% vs 4.3%, p =0.01, 7.3% vs 2.9%, p = 0.01, respectively). DEA identified 1552 genes (2.7% of total genes) with increased expression and 3344 with decreased expression (5.4% of total genes) in EMD when compared to non-EMD. Additionally, EMD cases were enriched for the CD-1, PR, and CTA myeloma gene expression signatures.
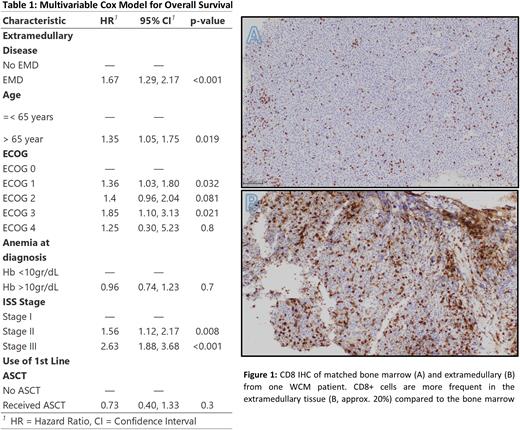
In the WCM cohort, EMD was also associated with worse OS relative to non-EMD (HR 2.7, 95% CI 1.1 - 6.5, p= 0.02). Visual assessment of IHC revealed heterogeneity in frequency and spatial distribution of various immune cell subsets between the matched BM and EMD samples, with striking enrichment of CD8+ and FOXP3+ cells observed in the EMD of one patient (Image 1). In order to characterize the spatial relationship, cellular interactions, and genomic characteristics in these samples, mIHC, WES and RNAseq studies are being performed on the matched samples.
Discussion Consistent with previous studies, patients with EMD during their disease course have a lower M-protein, and higher Hb at diagnosis of MM, and experience shorter mOS and PFS when compared to non-EMD patients. We observed no differences in the frequencies of recurrent translocations or copy number changes between these groups. EMD cases were enriched for TP53 mutations and bi-allelic TP53 inactivation, both of which are prognostic. There are differences in the gene expression profile of EMD patients in the BM at time of diagnosis compared to non-EMD, including enrichment for CD-1, PR, and CTA signatures, the latter two of which have been associated with poor outcomes. MxIF, WES, and RNAseq of BM and EMD samples is currently ongoing in order to further explore the tumor microenvironment and genomic landscape associated with EMD
Disclosures
Monge:Bristol Myers Squibb: Consultancy. Niesvizky:Takeda: Consultancy, Research Funding; Karyopharm: Consultancy, Research Funding; Janssen: Consultancy, Research Funding; GlaxoSmithKline: Consultancy, Research Funding; Bristol-Myers Squibb: Consultancy, Research Funding. Bustoros:Bristol Myers Squibb: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Takeda: Consultancy, Honoraria; Meniarini: Consultancy, Honoraria; Epizyme: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.